Artificial Intelligence in Drug Discovery
 Date: June 2020
Date: June 2020
Prepared by SIC Member: Lorraine V. Kalia, MD, PhD
Authors: Igor Jurisica, PhD, DSc; Alix Lacoste, PhD
Editor: Un Jung Kang, MD
Artificial Intelligence (AI) encompasses the theory, development, and application of computer systems that can learn, improve performance, and perform tasks associated with human intelligence, such as playing chess or Go, recognizing complex patterns in diverse data including images and sequences of strings, recognizing speech and language, and supporting complex decision-making processes. This human-like decision-making by computers is already starting to transform how we diagnose human disease (e.g., 1,2). Equally (or more) exciting is the potential of applying AI with ‘big data’ to the field of drug discovery, especially for neurological disorders without disease-modifying therapies such as Parkinson’s disease. Here we asked two experts in the field of AI, one from academia (Dr. Jurisica) and one from industry (Dr. Lacoste), to discuss the role they see for AI in drug discovery.
How can AI be applied to drug discovery?
Dr. Jurisica
It is essential to recognize that overall success of the application of AI depends on the algorithm, the data, and the process – underestimating any of these three components would result in a suboptimal solution at best, and complete failure at worst. Drug discovery uses diverse data, robotic screening, comprehensive pattern discovery, process optimization, and multi-faceted decision making, organized into rich workflows and pipelines. There are multiple opportunities for diverse AI algorithms to aid and improve many of these steps.
The drug discovery pipeline starts with identifying possible targets of interest by analysing rich, multi-omic datasets and finding useful non-trivial patterns – biomarkers and possible drug targets. As the number of assets decreases and cost raises, the pipeline covers high-throughput screening for possible molecules, lead identification and optimization, preclinical testing, clinical trials, and FDA approval (Figure 1). AI and integrative computational biology analyses have been applied to virtually all these steps – from identifying diagnostic/prognostic/predictive biomarkers and possible drug(able) targets, through automating screening and optimizing molecules, to guiding clinical trials and patient selection, and even helping to write patents for successful drugs. Supervised and unsupervised machine learning, text and image processing, text synthesis, and robotic automation with computational image characterization comprise the rich “AI toolbox” of algorithms for the drug discovery pipeline.
Increasingly, richer biomedical data and more comprehensive analyses and modeling are getting closer to achieving true precision or personalized medicine. We have a progressively improved understanding of how mutations affect protein interactions (3). In turn, this information is enabling us to predict how the interactome is rewired in individual patients, and how this affects drug mechanism of action, and thus patient response to treatment (4). Improved image analysis enables non-invasive monitoring of patient response to treatment and thus enhances clinical trial data.
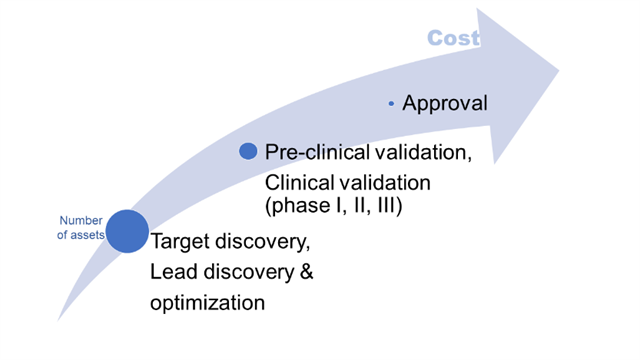
Figure 1. Large attrition rate and increasing costs with each step are the main challenges in the traditional drug discovery pipeline.
Dr. Lacoste
AI can enhance experts’ knowledge and ability to find insights in drug discovery in multiple ways. One is to intelligently digest and summarize the vast amount of scientific information that is published. Techniques such as natural language processing and text mining can identify biomedical terms and relationships between those terms, such as genes implicated in a disease, by scanning and interpreting the scientific literature. Information extracted from text can be combined with database information to allow scientists to amplify their knowledge surrounding a drug, gene, or disease. In addition to enhancing experts directly, aggregated information can be used by algorithms to suggest new areas of research. For example, target identification algorithms make use of known information to suggest new targets or pathways for a disease of interest. These algorithms come in different forms. Scientists can use rules on a knowledge network to identify genes that might be closely related to a disease of interest through common mechanisms with other genes for example. In other variations, experts can teach the system to learn by itself, typically based on known examples, and through machine learning methods. AI is also increasingly being applied to personalized medicine: identifying patient groups that might respond best to a specific treatment. In this case, AI works similarly by analyzing vast amounts of data, often both genetic and clinical, to propose patient groups and treatments. Finally, AI is being applied to designing better compounds more efficiently. AI can design compounds based on multi-parametric optimizations that factor in properties chemists seek to optimize for a molecule, for example, stability and binding affinity.
What are the current advantages and limitations of AI in drug discovery?
Dr. Jurisica
AI offers several significant advantages to the traditional drug discovery pipeline; for example, finding solutions that we would miss otherwise, providing richer annotation, and finding more solutions faster and thus cheaper compared to traditional strategies. There are many successful academic examples of using AI in drug discovery, and hundreds of AI companies work in this field. In 2019 Deep Knowledge Analytics compiled a list of the top 100 AI academic and industrial leaders in drug discovery and advanced healthcare (5).
To maximize the advantages of AI algorithms, it is useful to consider the full drug discovery workflow. AI alone would not get us far – while the AI toolbox includes useful approaches in machine learning, data mining, and knowledge representation, we need high quality data, rich ontologies and annotations, and many non-AI approaches, such as graph theory (6). Over the years, accuracy and speed of AI algorithms have improved tremendously, often thanks to advances in computing platforms. One such example includes a recent successful application of convolutional neural networks using GPU acceleration (7), which includes image analysis as well as bio-activity prediction in structure-based drug discovery (8).
Although progress has been made, the main challenge of many current applications includes limited transfer learning (i.e., moving from one application domain to another without re-training), and a system’s (in)ability to explain the solution. Explainable AI is being discussed, but many systems work as “black boxes” – they provide a solution, but do not explain “why” and “how” it was reached. There are many examples highlighting how some systems make a fairly correct decision or provide a reasonable solution, but for the wrong reasons. Carefully controlling characteristics and limiting biases of training data, and considering what features are extracted and used during learning is paramount. Especially in the medical field, we need full transparency of the reasoning and complete control of the training and validation processes. The system needs to also know its limitations, providing an “I do not know” answer rather than a probable solution that is incorrect. Ensuring we can estimate confidence on a given prediction, rather than just a cohort-based estimate is needed for individualized medicine.
While we have large volumes of molecular data on patients with specific diseases, we still have very limited information on patients themselves. Improving speed and accuracy of detecting non-invasive biomarkers, measuring microbiome and its changes, considering circadian rhythm and its effect on treatment, measuring information about life style, sleep, etc. from wearable devices will provide the necessary additional information for improving our understanding of why and how patients respond to therapy (9). Interplay of immune system, physical activity, hormones, and pharmaceuticals will need to be considered together to push treatment into a new level of personalization. Understanding how we age, how these processes relate to disease, and how we can control them will provide new targets for drug discovery (10). AI provides an essential component for all these diverse tasks.
Dr. Lacoste
AI is often limited by the quality, quantity, and availability of data. One of the main tools of AI, machine learning, ideally requires many training examples, both positive and negative to perform well. In fact, the machine learning field saw a big revival and improvement in technology following the release of a training set composed of hand drawn digits (MNIST). Machine learning algorithms learn by identifying patterns in the positive and negative examples. However, researchers typically look for drugs where there are few or none that are currently effective. For example, when searching to repurpose drugs for L-dopa induced dyskinesia (LID), my colleagues and I used a list of drugs that have demonstrated a reduction of LID in animal models as a proxy for what we were looking for: efficacy in humans (11). Negative examples are even more difficult to come by since the absence of positive results in an experiment does not actually mean that the object tested is negative; it could be that the experiment did not work or is not well-suited. In addition to issues with training algorithms, the dearth of positive and negative examples makes it difficult to evaluate whether an algorithm is predicting valuable data without running lengthy and costly experiments. This data limitation is one that AI faces especially in “discovery” fields, much more than in other fields where AI is typically applied, such as recommendation systems or image recognition.
Despite some of these challenges, the opportunities are great. As the field grows, we will develop more benchmark data sets that will enable bigger leaps. We have already seen some early successes where AI can accelerate discoveries in target identification or compound optimization, and more impressively, can recommend new avenues of research that experts would not have prioritized on their own. I am a firm believer that AI in drug discovery requires biomedical experts to be co-developers of the technologies to be successful (12).
Summary:
Pharmacological treatment of most movement disorders remains a huge challenge and thus there continues to be a major need for new therapies. As described above, the application of AI has the potential to expedite the traditional drug discovery pipeline, to reveal novel druggable targets and pathways, and eventually to facilitate delivery of true precision medicine. Examples of early work using AI for drug discovery in movement disorders include drug screening campaigns to identify drugs that may reduce levodopa-induced dyskinesia (11) or inhibit alpha-synuclein oligomers (13). Work to date is in its preliminary stages but it is anticipated that AI will help to change the treatment landscape for movement disorders. As in all areas of scientific discovery, the adage “garbage in, garbage out” holds true for AI driven research. Thus, to help make AI an effective tool for movement disorders drug discovery, we need to continue to support and collaborate on both collecting and sharing high quality data.
References:
1) Esteva A et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115-118.
2) Hollon TC et al. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat Med 2020;26:52-58.
3) IMEx Consortium Curators et al. Capturing variation impact on molecular interactions in the IMEx Consortium mutations data set. Nat Commun 2019;10:10.
4) Mandilaras V et al. TP53 mutations in high grade serous ovarian cancer and impact on clinical outcomes: a comparison of next generation sequencing and bioinformatics analyses. Int J Gynecol Cancer 2019 pii: ijgc-2018-000087.
5) www.ai-pharma.dka.global/ai-leaders/
6) Huang LC et al. Graph theory and network topological metrics may be the potential biomarker in Parkinson's disease. J Clin Neurosci 2019;68:235-242.
7) www.nvidia.com/en-us/deep-learning-ai/solutions/data-science/
8) www.atomwise.com/
9) Holzinger A et al. Why imaging data alone is not enough: AI-based integration of imaging, omics, and clinical data. Eur J Nucl Med Mol Imaging 2019;46:2722-2730.
10) www.bioagelabs.com
11) Johnston TH et al. Repurposing drugs to treat l-DOPA-induced dyskinesia in Parkinson's disease. Neuropharmacology 2019;147:11-27.
12) www.statnews.com/2019/11/01/ai-revolutionize-drug-discovery-experts-involved/
13) Maclagan LC et al. Identifying drugs with disease-modifying potential in Parkinson's disease using artificial intelligence and pharmacoepidemiology. Pharmacoepidemiol Drug Saf. 2020 May 14. doi: 10.1002/pds.5015






Submit Your Comment